
Heterozygote Peaks
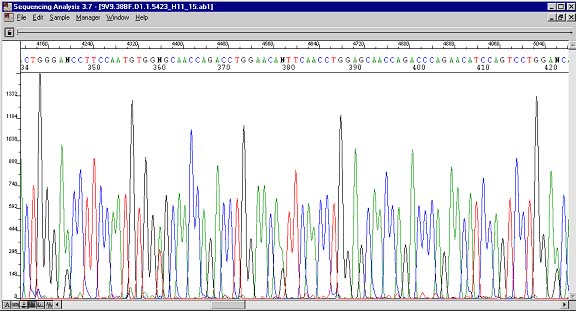
Sequence with four heterozygous (N) peaks.
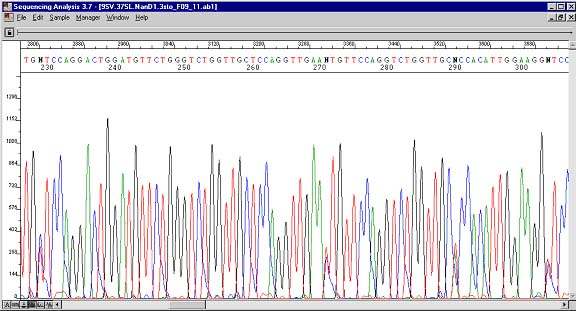
Reverse sequence showing the complement of the same four heterozygous (N) peaks.
Heterozygous peaks are displayed as at least two overlapping peaks in the same position. They are indicative of the presence of two species of DNA that differ at these locations. The reverse sequence shows that this in not an anomaly. Single nucleotide polymorphisms are found in this way. One important note of caution is that not all heterozygous peaks will be called an unknown (N). How unfortunate would it be if these went unnoticed during a site-directed mutagenesis experiment! If heterozygous peaks arise unexpectedly, then pause should be taken to rectify the problem. There could be mutagen contamination in your PCR reagents. To be absolutely certain that a base is heterozygous, one could design two primers with 3' ends at the heterozygous base and see in which reaction PCR product results.
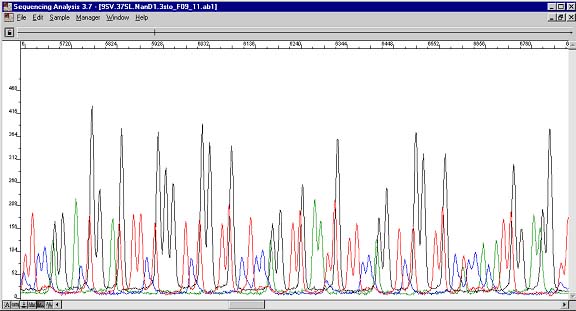
Raw data view of reverse sequence. Raw data view is available through ABI Prism Sequencing Analysis. The peak height is roughly half the height of other peaks of the same color. This should not be used as an accurate way to quantify proportions of DNA species.
