
Multiple Peaks
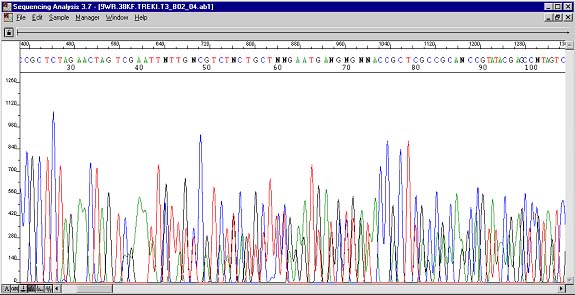
Multiple peaks describe the chromatogram peak characteristic that occurs when two types of dye terminated fragments are the same size. Or in other words, when two sequences are contained in the same reaction. The same result would occur if two sequencing reactions were simply mixed together. Multiple peak data comes in many varieties and often one data set is weaker that the other. A typical example is shown above where a vector primer encounters two different sequences at the restriction site. In some of the plasmid molecules the insert may not be present, be inverted, or include other digest products. Do not pool plamid preps. The above example could also be the result of a primer that anneals to a repeat region. Multiple peaks can also be present from the beginning of a read. In both PCR and plasmid templates these reasons are the presence of other binding sites for the primer on the same template, or primer contamination with another primer. Be sure that all of the unincorporated amplification primers and nonspecific PCR products are absent from a PCR template sample before submitting for sequencing. It would be to your benefit to visualize your template on a quick agarose gel to confirm the presence of one product and one product only.
