
Primer Dimers
Primer dimers occur when primers are accidentally designed with inverted regions. This allows them to anneal to other primer molecules and, in effect, become linear mini-templates.
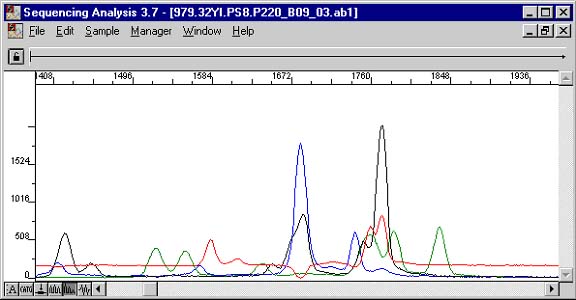
A measure of the strength or spontaneity of this dimerizing is the reaction's disassociation constant (DG). The value is found on most primer design programs and should not be less than -3.5 kcal/mol (or more negative than, in other words). If they are, the primers will favor dimerizing more than annealing to their true target. The example below really illustrates this point well. Fortunately for this purpose, a 5' overhang in one of the conformations was long enough to allow its sequencing products to be included in the filtrate. Not all conformations will contribute to detected signal, but they will all be sequestered from the expected reaction. The two CCGG sections really compound the problem. You can actually read the complement of the overhang in the smaller first bases of the chromatogram. Most of the primers were unterminated upon the short extension and account for the larger fragments and their taller peaks in the messier section as they themselves dimerized.
Most primer dimers result in a failed chromatogram since their short sequencing products (from small 5' overhangs) remain in the retentate. These can only be diagnosed using a primer design program such as the ones listed below.
Primer design programs should always be used before sending the primer sequence in for synthesis. Raising the annealing temperature to be more stringent during a resequence will not work; redesigning the primer is necessary.
Primer Design Websites:
- http://www.autoprimer.com/ Free 30 day trial, free to Beckman Coulter customers.
- http://www.idtdna.com/
- OligoAnalyzer3.0 - Tm, hairpins, dimers, %GC
- PrimerQuest - primer design, minimum template size is 200bp
- Basic Sciences at the Northwestern University Medical School - oligonucleotide properties calculator. http://www.basic.nwu.edu/biotools/oligocalc.html
- http://healthcare.partners.org/dnacore/oligotoolform.cfm - Takes into account degererate bases. Do not use degenerate primers in SNP detection.
Sequencing Data
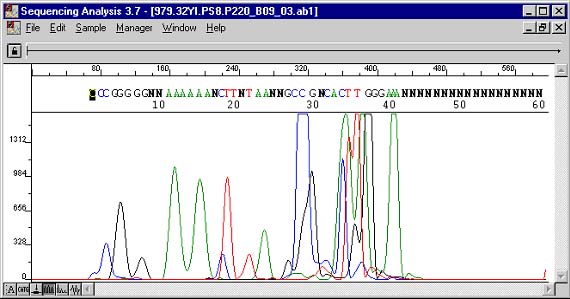
Delta G -9.75 kcal/mole
Base Pairs 4
5' CCTAAGTTCCGGCCGGAGAGACGTCAGCACACCGG |||| | | |||| 3' GGCCACACGACTGCAGAGAGGCCGGCCTTGAATCC
Delta G -9.75 kcal/mole
Base Pairs 4
5' CCTAAGTTCCGGCCGGAGAGACGTCAGCACACCGG |||| | | | | | | |||| 3' GGCCACACGACTGCAGAGAGGCCGGCCTTGAATCC
Delta G -9.75 kcal/mole
Base Pairs 4
5' CCTAAGTTCCGGCCGGAGAGACGTCAGCACACCGG |||| 3' GGCCACACGACTGCAGAGAGGCCGGCCTTGAATCC
Delta G -9.45 kcal/mole
Base Pairs 6
5' CCTAAGTTCCGGCCGGAGAGACGTCAGCACACCGG | | | |||||| | | | 3' GGCCACACGACTGCAGAGAGGCCGGCCTTGAATCC
