
Secondary Structure
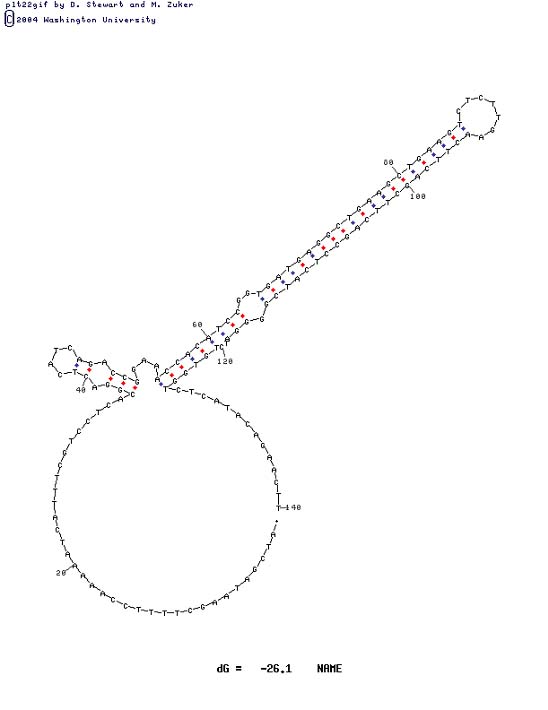
This is one of several possible conformations for the 140 bases surrounding the problem area as attained from http://biotools.idtdna.com/mFold/ A major secondary structure motif will be obvious in all conformations.
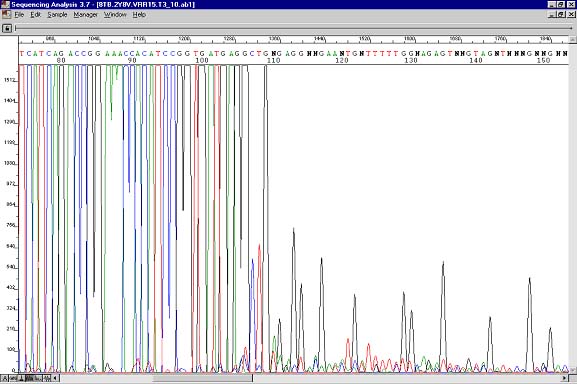
Sequencing result where data abruptly stops. The polymerase enzymes could not get through secondary structure occurring at this spot.
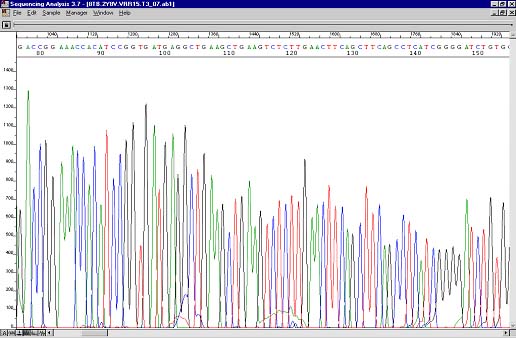
Resequencing result where the majority of the polymerase enzymes sequenced through the hairpin. A small drop in peak height can be observed in the problem area.
DNA secondary structure is a common occurrence. It plays a role in splice variants and can be at intron/exon interfaces. Hairpins are engineered into siRNA and RNAi constructs while complex secondary structure knots containing triplex interactions between bases occur in nature. While these work wonderfully for silencing and interfering with a genetic target, or for switching between two genes that reside in different reading frames in the same location in some viruses, secondary structure is detrimental to PCR, and certainly PCR-based sequencing. The local high melting temperature of these problem areas and direct hairpin-polymerase interactions are to blame. To overcome these obstacles we use an in house sequencing brew affectionately dubbed "The Works", both because it uses every compatible chemical and enzyme we stock and because it works. The cocktail is as follows: ABI dGTPv3.0 Terminator, 1l (sequencing kit); ABI BigDye Terminator v3.1, 2l (sequencing kit); ABI BigDye Terminator v1.1, v3.1 5X Sequencing Buffer, 2.2l (or enough to bring the final concentration to 1X considering that each l of sequencing kit contributes 2.5X); 5% DMSO, final (Sigma); 1M Betaine, final (Sigma); 600ng plasmid template or 100ng linear PCR DNA; and 6pmol primer. Like most sequencing problems, secondary structure data have a range of results. These can be in the form of a small, unobtrusive drop in peak height, to a dead stop. But stranger things have happened, as illustrated below.
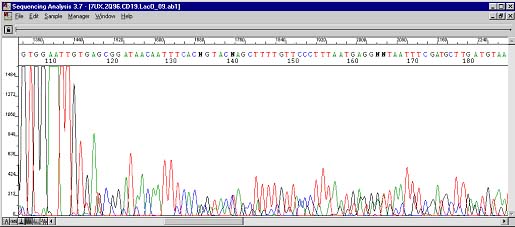
The above case was very interesting. Here was a plasmid containing an exon/intron interface. After the problem area two peak traces can be observed. Troubleshooting with the customer ruled out the possibility of two types of inserts in the vector. This case was actually the impetus for the creation of The Works. Pay attention to the four Ts at position 145 and the green (A), black (G), blue (C), and blue (C) peaks directly below.
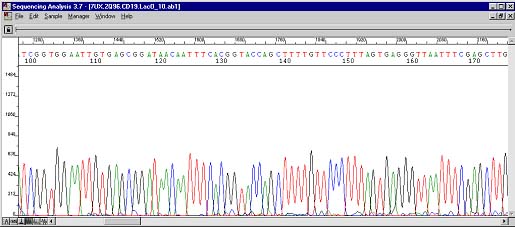
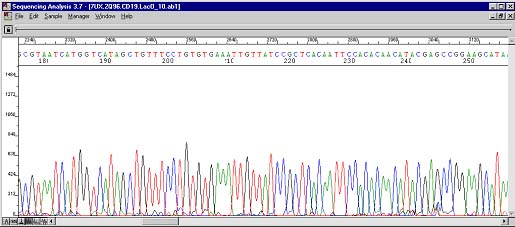
These are the resequencing results. The four Ts can still be seen at roughly the same position albeit cleaner and more uniform. The underlying peaks now show up at base position 245. Previously during PCR, the polymerases were doing three things: most were stopping at 115 (hence the big drop), some were sequencing through the secondary structure (the four Ts), and fewer still were actually skipping the structure altogether (the AGCC occurring 100 bases too early)!
